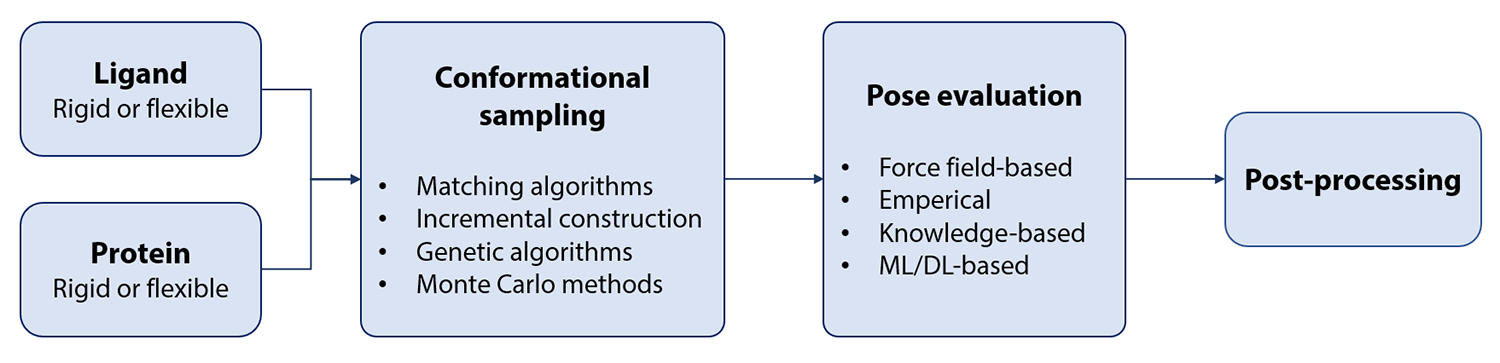
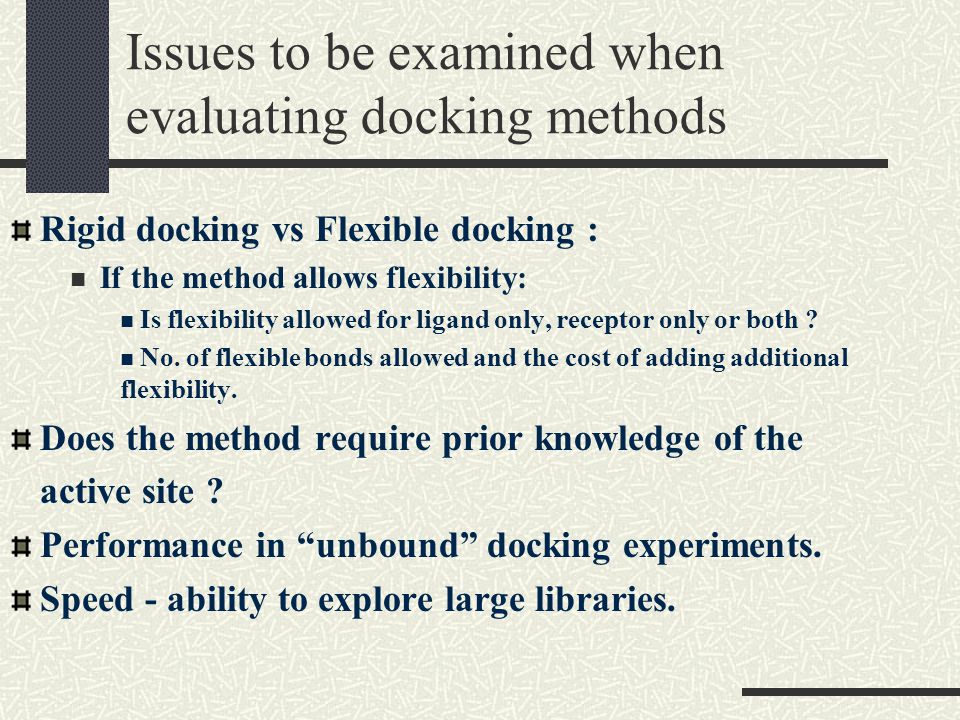
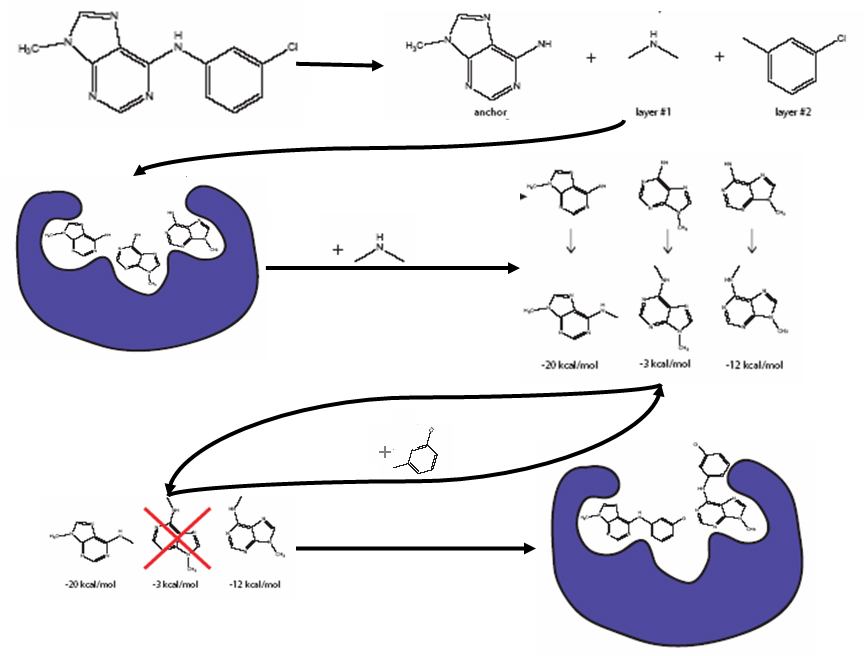
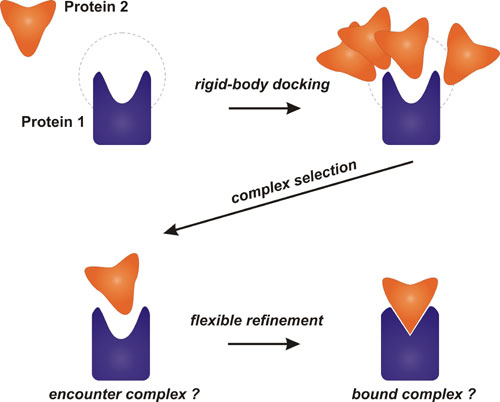
Deep Learning Model for Efficient Protein–Ligand Docking with Implicit Side-Chain Flexibility | Journal of Chemical Information and Modeling
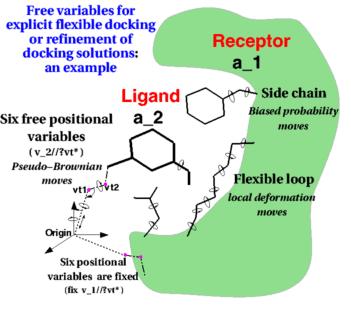
Flexible Docking of Biological Macromolecules - Theoretical Biophysics (T38) - Biomolecular Dynamics
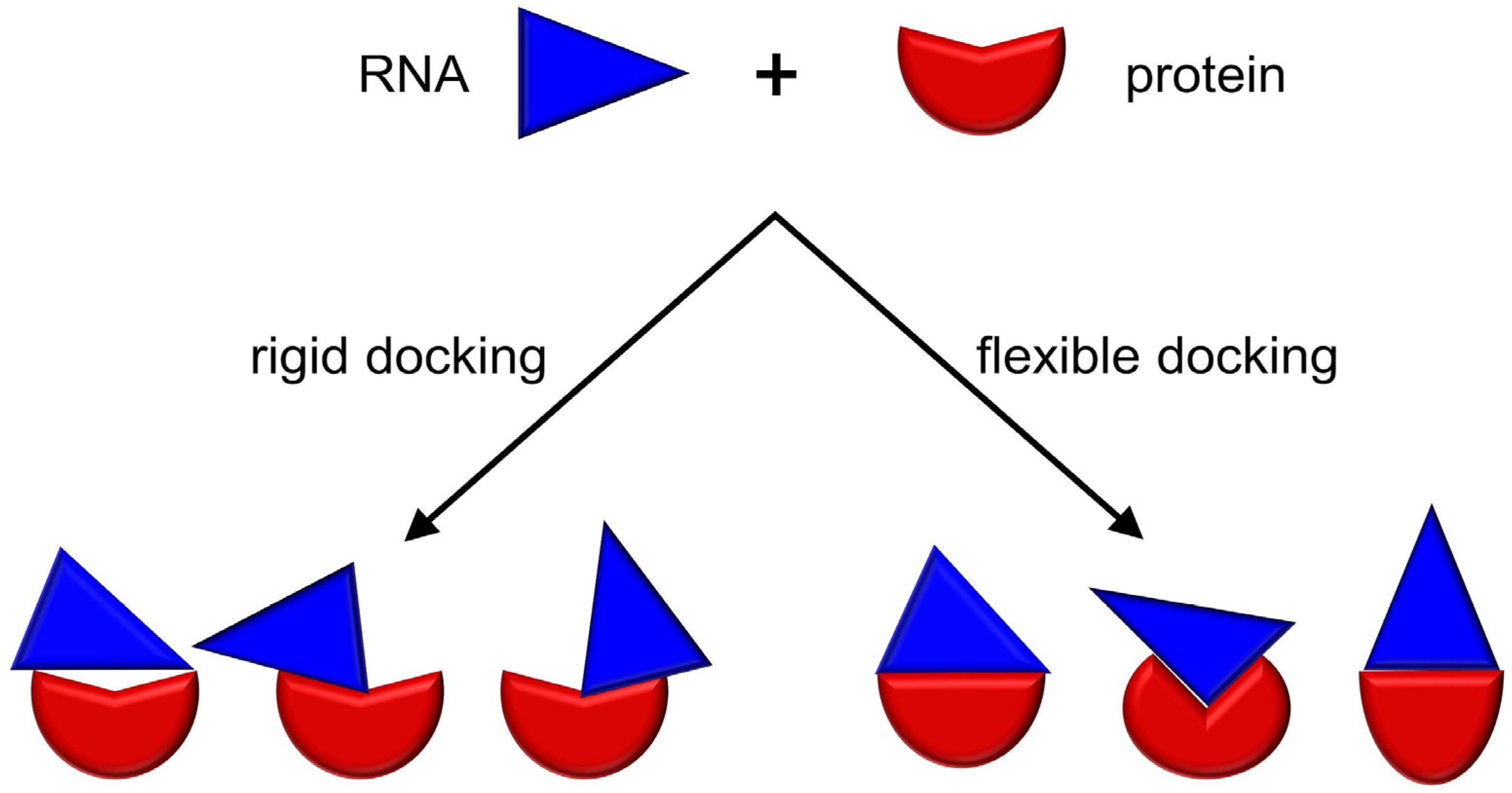
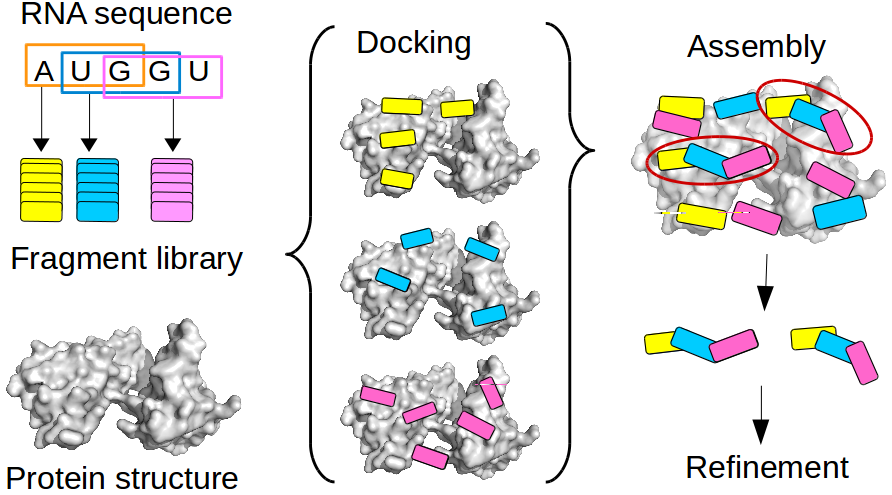
Genes | Free Full-Text | Bioinformatics Tools and Benchmarks for Computational Docking and 3D Structure Prediction of RNA-Protein Complexes

Combining classical molecular docking with self-consistent charge density-functional tight-binding computations for the efficient and quality prediction of ligand binding structure - Amar Y Al-Ansi, Haorui Lu, Zijing Lin, 2022
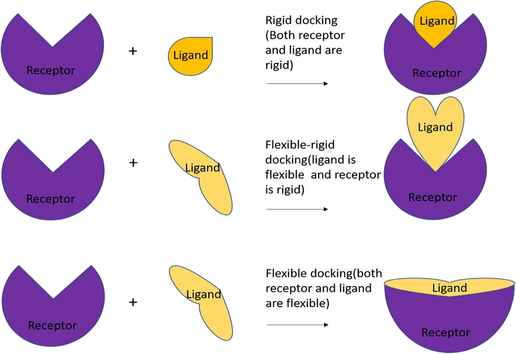
Molecular docking in organic, inorganic, and hybrid systems: a tutorial review | Monatshefte für Chemie - Chemical Monthly

Molecules | Free Full-Text | How 'Protein-Docking' Translates into the New Emerging Field of Docking Small Molecules to Nucleic Acids?